West Nile Virus Prediction

A simple approach with focus on thought process when working on a data set. For aspiring data analyst/scientist.
For this project, I took the Kaggle competition data set to perform my analysis. As I want to validate my prediction, I took out a hold out set from the train data set, instead of using the test data set provided.
Background
In 2002, the first human cases of West Nile virus were reported in Chicago. By 2004 the City of Chicago and the Chicago Department of Public Health (CDPH) had established a comprehensive surveillance and control program that is still in effect today.
Every week from late spring through the fall, mosquitos in traps across the city are tested for the virus. The results of these tests influence when and where the city will spray airborne pesticides to control adult mosquito populations.
Given weather, location, testing, and spraying data, the idea is to predict when and where different species of mosquitos will test positive for West Nile virus. A more accurate method of predicting outbreaks of West Nile virus in mosquitos will help the City of Chicago and CPHD more efficiently and effectively allocate resources towards preventing transmission of this potentially deadly virus.
Problem Statement
With the given data, predict if a species of mosquito carries the West Nile virus.
Approach
In this case study, we are looking to predict if a mosquito carries the West Nile virus. A supervised classification model will be use to do the prediction.
Success Metrics
I would use recall as my success metrics instead of precision, the reason being that you want to predict which mosquito indeed has the virus so that focus can be placed on those estate with high rate of mosquito carrying the virus.
Recall: Out of all the True Condition Positive, how many did the model predict correctly. Precision: Out of all the Predicted Condition Positive, how many did the predict correctly.
# somehow.. loading in the packages feel like the process Bae goes through in her make up routine~~~
# standard packages (the cleansing)
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import sqlite3
from collections import Counter
# preparation of data for modeling(foundation, moisturizing, concealer, blah blah..)
from sklearn.preprocessing import MultiLabelBinarizer
from sklearn.model_selection import train_test_split
from imblearn.combine import SMOTETomek
from sklearn.preprocessing import StandardScaler
# data modeling (putting on all the coloring & blushes)
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import GridSearchCV
# data validation (asking me how she look~ that's the stage where I usually died~ [-_-"])
from sklearn.model_selection import cross_val_score
from sklearn.metrics import classification_report, recall_score, confusion_matrix, roc_auc_score, roc_curve
# tree visualisation
from sklearn.tree import export_graphviz
from sklearn.externals.six import StringIO
from IPython.display import Image
import pydot
%matplotlib inline
Understanding the Dataset
It’s attempting to jump right into cleaning the data set and get the heavy lifting out of the way, but I am going to hold my horses a bit, and have an overview of the data set that I am working on. I could have done everything within Python itself, but since I want to keep my SQL sharp… I am going to do another step by loading the CSV into SQLite & perform my ETL there.
# set up the sql database
sqlite_db = 'Dataset/westnile.sqlite'
conn = sqlite3.connect(sqlite_db)
c = conn.cursor()
# load in the dataset
csv_spray = 'Dataset/spray.csv'
csv_weather = 'Dataset/weather.csv'
csv_data = 'Dataset/train.csv'
# taking a look at the data set I am dealing with
spray = pd.read_csv(csv_spray)
spray.head()
| Date | Time | Latitude | Longitude |
|---|---|---|---|
| 2011-08-29 | 6:56:58 PM | 42.391623 | -88.089163 |
| 2011-08-29 | 6:57:08 PM | 42.391348 | -88.089163 |
| 2011-08-29 | 6:57:18 PM | 42.391022 | -88.089157 |
| 2011-08-29 | 6:57:28 PM | 42.390637 | -88.089158 |
| 2011-08-29 | 6:57:38 PM | 42.390410 | -88.088858 |
weather = pd.read_csv(csv_weather)
weather.head()
| Station | Date | Tmax | Tmin | Tavg | Depart | DewPoint | WetBulb | Heat | Cool | … | CodeSum | Depth | Water1 | SnowFall | PrecipTotal | StnPressure | SeaLevel | ResultSpeed | ResultDir | AvgSpeed |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2007-05-01 | 83 | 50 | 67 | 14 | 51 | 56 | 0 | 2 | … | 0 | M | 0.0 | 0.00 | 29.10 | 29.82 | 1.7 | 27 | 9.2 | |
| 2 | 2007-05-01 | 84 | 52 | 68 | M | 51 | 57 | 0 | 3 | … | M | M | M | 0.00 | 29.18 | 29.82 | 2.7 | 25 | 9.6 | |
| 1 | 2007-05-02 | 59 | 42 | 51 | -3 | 42 | 47 | 14 | 0 | … | BR | 0 | M | 0.0 | 0.00 | 29.38 | 30.09 | 13.0 | 4 | 13.4 |
| 2 | 2007-05-02 | 60 | 43 | 52 | M | 42 | 47 | 13 | 0 | … | BR HZ | M | M | M | 0.00 | 29.44 | 30.08 | 13.3 | 2 | 13.4 |
| 1 | 2007-05-03 | 66 | 46 | 56 | 2 | 40 | 48 | 9 | 0 | … | 0 | M | 0.0 | 0.00 | 29.39 | 30.12 | 11.7 | 7 | 11.9 |
data = pd.read_csv(csv_data)
data.head()
| Date | Address | Species | Block | Street | Trap | AddressNumberAndStreet | Latitude | Longitude | AddressAccuracy | NumMosquitos | WnvPresent |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2007-05-29 | 4100 North Oak Park Avenue, Chicago, IL 60634,… | CULEX PIPIENS/RESTUANS | 41 | N OAK PARK AVE | T002 | 4100 N OAK PARK AVE, Chicago, IL | 41.954690 | -87.800991 | 9 | 1 | 0 |
| 2007-05-29 | 4100 North Oak Park Avenue, Chicago, IL 60634,… | CULEX RESTUANS | 41 | N OAK PARK AVE | T002 | 4100 N OAK PARK AVE, Chicago, IL | 41.954690 | -87.800991 | 9 | 1 | 0 |
| 2007-05-29 | 6200 North Mandell Avenue, Chicago, IL 60646, USA | CULEX RESTUANS | 62 | N MANDELL AVE | T007 | 6200 N MANDELL AVE, Chicago, IL | 41.994991 | -87.769279 | 9 | 1 | 0 |
| 2007-05-29 | 7900 West Foster Avenue, Chicago, IL 60656, USA | CULEX PIPIENS/RESTUANS | 79 | W FOSTER AVE | T015 | 7900 W FOSTER AVE, Chicago, IL | 41.974089 | -87.824812 | 8 | 1 | 0 |
| 2007-05-29 | 7900 West Foster Avenue, Chicago, IL 60656, USA | CULEX RESTUANS | 79 | W FOSTER AVE | T015 | 7900 W FOSTER AVE, Chicago, IL | 41.974089 | -87.824812 | 8 | 4 | 0 |
# OK~ looks like there is a bit of cleaning up to do. (Null values, weird values..) Better get ready my toilet brush.
Spray
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 14835 entries, 0 to 14834
Data columns (total 4 columns):
Date 14835 non-null object
Time 14251 non-null object
Latitude 14835 non-null float64
Longitude 14835 non-null float64
dtypes: float64(2), object(2)
memory usage: 463.7+ KB
None
--------------------
Weather
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 2944 entries, 0 to 2943
Data columns (total 22 columns):
Station 2944 non-null int64
Date 2944 non-null object
Tmax 2944 non-null int64
Tmin 2944 non-null int64
Tavg 2944 non-null object
Depart 2944 non-null object
DewPoint 2944 non-null int64
WetBulb 2944 non-null object
Heat 2944 non-null object
Cool 2944 non-null object
Sunrise 2944 non-null object
Sunset 2944 non-null object
CodeSum 2944 non-null object
Depth 2944 non-null object
Water1 2944 non-null object
SnowFall 2944 non-null object
PrecipTotal 2944 non-null object
StnPressure 2944 non-null object
SeaLevel 2944 non-null object
ResultSpeed 2944 non-null float64
ResultDir 2944 non-null int64
AvgSpeed 2944 non-null object
dtypes: float64(1), int64(5), object(16)
memory usage: 506.1+ KB
None
--------------------
Data
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 10506 entries, 0 to 10505
Data columns (total 12 columns):
Date 10506 non-null object
Address 10506 non-null object
Species 10506 non-null object
Block 10506 non-null int64
Street 10506 non-null object
Trap 10506 non-null object
AddressNumberAndStreet 10506 non-null object
Latitude 10506 non-null float64
Longitude 10506 non-null float64
AddressAccuracy 10506 non-null int64
NumMosquitos 10506 non-null int64
WnvPresent 10506 non-null int64
dtypes: float64(2), int64(4), object(6)
memory usage: 985.0+ KB
None
Data Preparation (Cleaning & EDA)
Well, like that they say, data preparation takes up 80% of the data scientist work. Bam! It’s hard work. #nojoke
# convert Date into a date format
spray['Date'] = pd.to_datetime(spray['Date'], format='%Y/%m/%d')
weather['Date'] = pd.to_datetime(weather['Date'], format='%Y/%m/%d')
data['Date'] = pd.to_datetime(data['Date'], format='%Y/%m/%d')
# clean up the Average Temperature by re-calculating it
weather['Tavg'] = np.mean([weather.Tmax, weather.Tmin], axis=0)
# Interpolate the wetbulb
weather['WetBulb'] = weather.WetBulb.replace('M', np.nan)
weather['WetBulb'] = weather['WetBulb'].astype('float')
weather['WetBulb'] = weather['WetBulb'].interpolate()
# replace empty cell in CodeSum with 'Moderate'
weather['CodeSum'] = weather.CodeSum.replace(' ', 'Moderate')
# Interpolate the PrecipTotal
weather['PrecipTotal'] = weather.PrecipTotal.replace('M', np.nan)
weather['PrecipTotal'] = weather.PrecipTotal.replace(' T', np.nan)
weather['PrecipTotal'] = weather['PrecipTotal'].astype('float')
weather['PrecipTotal'] = weather['PrecipTotal'].interpolate()
# Interpolate the StnPressure
weather['StnPressure'] = weather.StnPressure.replace('M', np.nan)
weather['StnPressure'] = weather['StnPressure'].astype('float')
weather['StnPressure'] = weather['StnPressure'].interpolate()
# use mean for missing sealevel
weather['SeaLevel'] = weather.SeaLevel.replace('M', np.nan)
weather['SeaLevel'] = weather['SeaLevel'].astype('float')
weather.SeaLevel.fillna(weather.SeaLevel.mean(), inplace=True)
weather['SeaLevel'] = round(weather.SeaLevel, 2)
# use mean for missing average wind speed
weather['AvgSpeed'] = weather.AvgSpeed.replace('M', np.nan)
weather['AvgSpeed'] = weather['AvgSpeed'].astype('float')
weather.AvgSpeed.fillna(weather.AvgSpeed.mean(), inplace=True)
weather['AvgSpeed'] = round(weather.AvgSpeed, 1)
# Replace other Missing and Trace with -1
weather = weather.replace('M', -1)
weather = weather.replace('T', -1)
weather = weather.replace(' T', -1)
weather = weather.replace(' T', -1)
# checking on the period of the data
def show_date(df):
date = list(df.map(lambda x: x.to_period('m')).unique())
print(sorted(date))
# let's take a look at the the period of the data set
Spray Date (Year)
[Period('2011-08', 'M'), Period('2011-09', 'M'), Period('2013-07', 'M'), Period('2013-08', 'M'), Period('2013-09', 'M')]
None
====================
Weather Date(Year)
[Period('2007-05', 'M'), Period('2007-06', 'M'), Period('2007-07', 'M'), Period('2007-08', 'M'), Period('2007-09', 'M'), Period('2007-10', 'M'), Period('2008-05', 'M'), Period('2008-06', 'M'), Period('2008-07', 'M'), Period('2008-08', 'M'), Period('2008-09', 'M'), Period('2008-10', 'M'), Period('2009-05', 'M'), Period('2009-06', 'M'), Period('2009-07', 'M'), Period('2009-08', 'M'), Period('2009-09', 'M'), Period('2009-10', 'M'), Period('2010-05', 'M'), Period('2010-06', 'M'), Period('2010-07', 'M'), Period('2010-08', 'M'), Period('2010-09', 'M'), Period('2010-10', 'M'), Period('2011-05', 'M'), Period('2011-06', 'M'), Period('2011-07', 'M'), Period('2011-08', 'M'), Period('2011-09', 'M'), Period('2011-10', 'M'), Period('2012-05', 'M'), Period('2012-06', 'M'), Period('2012-07', 'M'), Period('2012-08', 'M'), Period('2012-09', 'M'), Period('2012-10', 'M'), Period('2013-05', 'M'), Period('2013-06', 'M'), Period('2013-07', 'M'), Period('2013-08', 'M'), Period('2013-09', 'M'), Period('2013-10', 'M'), Period('2014-05', 'M'), Period('2014-06', 'M'), Period('2014-07', 'M'), Period('2014-08', 'M'), Period('2014-09', 'M'), Period('2014-10', 'M')]
None
====================
[Period('2007-05', 'M'), Period('2007-06', 'M'), Period('2007-07', 'M'), Period('2007-08', 'M'), Period('2007-09', 'M'), Period('2007-10', 'M'), Period('2009-05', 'M'), Period('2009-06', 'M'), Period('2009-07', 'M'), Period('2009-08', 'M'), Period('2009-09', 'M'), Period('2009-10', 'M'), Period('2011-06', 'M'), Period('2011-07', 'M'), Period('2011-08', 'M'), Period('2011-09', 'M'), Period('2013-06', 'M'), Period('2013-07', 'M'), Period('2013-08', 'M'), Period('2013-09', 'M')]
None
From the looks of the period of the data, there is only record of:
- spray on 2011 (Aug - Sep) & 2013 (Jul - Sep)
- weather on 2007 to 2014 (May - Oct)
- Data on 2007 (May - Oct), 2009 (May - Oct), 2011 (Jun - Sep), 2013 (Jun - Sep)
I would expect there will be rows with quite a bit of missing values that when the tables are combined.
data.to_sql('data', con=conn, if_exists='replace', index=False)
spray.to_sql('spray', con=conn, if_exists='replace', index=False)
weather.to_sql('weather', con=conn, if_exists='replace', index=False)
# time to join the tables together and see the combined output
sql_query = '''
SELECT t."Date", t."Address", t."Species", t."Block", t."Street", t."Trap", t."AddressNumberAndStreet",
t."Latitude", t."Longitude",t."AddressAccuracy", t."NumMosquitos", t."WnvPresent",
CASE WHEN s."Latitude" IS NOT NULL THEN 1 END AS 'With_spray',
w."Station", w."Tmax", w."Tmin", w."Tavg", w."Depart", w."DewPoint",w."WetBulb", w."Heat", w."Cool", w."Sunrise",
w."Sunset", w."CodeSum", w."Depth", w."Water1", w."SnowFall", w."PrecipTotal", w."StnPressure",w."SeaLevel",
w."ResultSpeed", w."ResultDir", w."AvgSpeed"
FROM data t
LEFT JOIN spray s on ROUND(s."Latitude",3) = ROUND(t."Latitude",3) AND ROUND(s."Longitude",3) = ROUND(t."Longitude",3) AND s."Date" = t."Date"
LEFT JOIN weather w on w."Date" = t."Date"
'''
combined_data = pd.read_sql(sql_query, con=conn)
combined_data.head()
| Date | Address | Species | Block | Street | Trap | AddressNumberAndStreet | Latitude | Longitude | AddressAccuracy | … | CodeSum | Depth | Water1 | SnowFall | PrecipTotal | StnPressure | SeaLevel | ResultSpeed | ResultDir | AvgSpeed |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2007-05-29 00:00:00 | 4100 North Oak Park Avenue, Chicago, IL 60634,… | CULEX PIPIENS/RESTUANS | 41 | N OAK PARK AVE | T002 | 4100 N OAK PARK AVE, Chicago, IL | 41.954690 | -87.800991 | 9 | … | BR HZ | 0 | -1 | 0.0 | 0.0 | 29.39 | 30.11 | 5.8 | 18 | 6.5 |
| 2007-05-29 00:00:00 | 4100 North Oak Park Avenue, Chicago, IL 60634,… | CULEX PIPIENS/RESTUANS | 41 | N OAK PARK AVE | T002 | 4100 N OAK PARK AVE, Chicago, IL | 41.954690 | -87.800991 | 9 | … | BR HZ | -1 | -1 | -1 | 0.0 | 29.44 | 30.09 | 5.8 | 16 | 7.4 |
| 2007-05-29 00:00:00 | 4100 North Oak Park Avenue, Chicago, IL 60634,… | CULEX RESTUANS | 41 | N OAK PARK AVE | T002 | 4100 N OAK PARK AVE, Chicago, IL | 41.954690 | -87.800991 | 9 | … | BR HZ | 0 | -1 | 0.0 | 0.0 | 29.39 | 30.11 | 5.8 | 18 | 6.5 |
| 2007-05-29 00:00:00 | 4100 North Oak Park Avenue, Chicago, IL 60634,… | CULEX RESTUANS | 41 | N OAK PARK AVE | T002 | 4100 N OAK PARK AVE, Chicago, IL | 41.954690 | -87.800991 | 9 | … | BR HZ | -1 | -1 | -1 | 0.0 | 29.44 | 30.09 | 5.8 | 16 | 7.4 |
| 2007-05-29 00:00:00 | 6200 North Mandell Avenue, Chicago, IL 60646, USA | CULEX RESTUANS | 62 | N MANDELL AVE | T007 | 6200 N MANDELL AVE, Chicago, IL | 41.994991 | -87.769279 | 9 | … | BR HZ | 0 | -1 | 0.0 | 0.0 | 29.39 | 30.11 | 5.8 | 18 | 6.5 |
# .info() of the combined table
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 21020 entries, 0 to 21019
Data columns (total 34 columns):
Date 21020 non-null object
Address 21020 non-null object
Species 21020 non-null object
Block 21020 non-null int64
Street 21020 non-null object
Trap 21020 non-null object
AddressNumberAndStreet 21020 non-null object
Latitude 21020 non-null float64
Longitude 21020 non-null float64
AddressAccuracy 21020 non-null int64
NumMosquitos 21020 non-null int64
WnvPresent 21020 non-null int64
With_spray 14 non-null float64
Station 21020 non-null int64
Tmax 21020 non-null int64
Tmin 21020 non-null int64
Tavg 21020 non-null float64
Depart 21020 non-null object
DewPoint 21020 non-null int64
WetBulb 21020 non-null float64
Heat 21020 non-null object
Cool 21020 non-null object
Sunrise 21020 non-null object
Sunset 21020 non-null object
CodeSum 21020 non-null object
Depth 21020 non-null object
Water1 21020 non-null int64
SnowFall 21020 non-null object
PrecipTotal 21020 non-null float64
StnPressure 21020 non-null float64
SeaLevel 21020 non-null float64
ResultSpeed 21020 non-null float64
ResultDir 21020 non-null int64
AvgSpeed 21020 non-null float64
dtypes: float64(10), int64(10), object(14)
memory usage: 5.5+ MB
# creating a new column of month
combined_data['Date'] = pd.to_datetime(combined_data['Date'], format='%Y/%m/%d')
combined_data['Month'] = combined_data.Date.dt.month
# get the MM-YY out from the 'Date' column
combined_data['mm'] = combined_data['Date'].map(lambda x: x.to_period('m'))
unique_date = list(combined_data['mm'].unique())
# round the longitude and latitude into 2 decimal
combined_data['Latitude'] = round(combined_data['Latitude'], 2)
combined_data['Longitude'] = round(combined_data['Longitude'], 2)
# Breaking up CodeSum to the various weather condition
combined_data['CodeSum'] = combined_data.CodeSum.apply(lambda c: c.split(' '))
mlb = MultiLabelBinarizer()
df_lab = mlb.fit_transform(combined_data['CodeSum'])
combined_data = combined_data.join(pd.DataFrame(df_lab, columns=mlb.classes_))
# Picture speaks a thousand words, one of my hack to visualise null values
plt.figure(figsize=(18,6))
sns.heatmap(combined_data.isnull(), yticklabels=False, cbar=False, cmap='plasma')
plt.show()
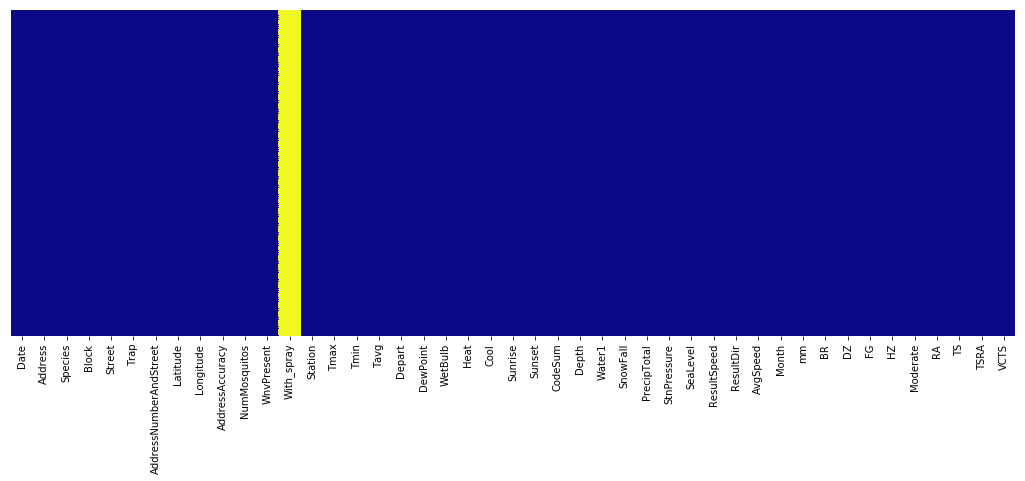
# see which mosquito tend to carry the virus
mos_wnv = combined_data[['Species', 'NumMosquitos', 'WnvPresent']].groupby(by='Species').sum()
mos_wnv.reset_index(inplace=True)
plt.figure(figsize=(20,6))
plt.bar(mos_wnv['Species'], mos_wnv['WnvPresent'])
plt.show()
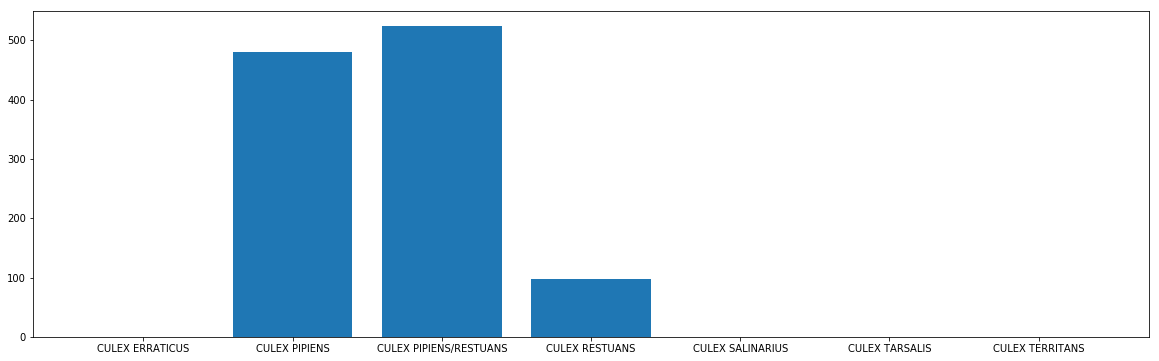
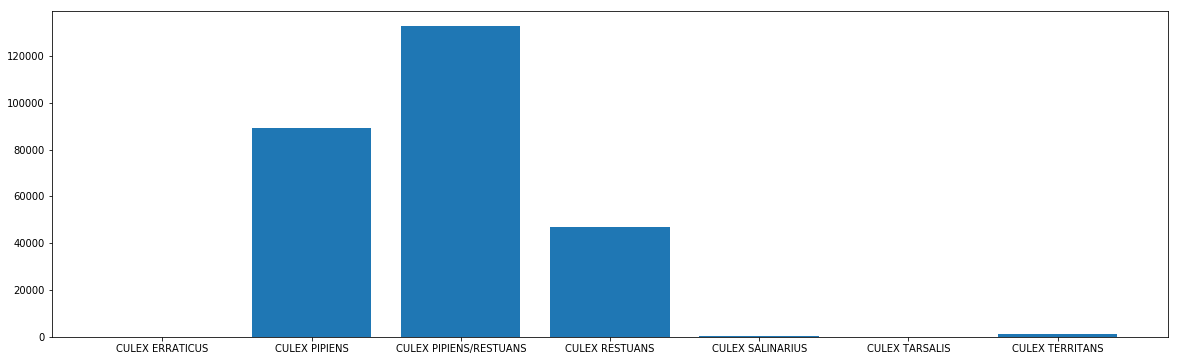
# see the number count of the mosquito
plt.figure(figsize=(20,6))
plt.bar(mos_wnv['Species'], mos_wnv['NumMosquitos'])
plt.show()
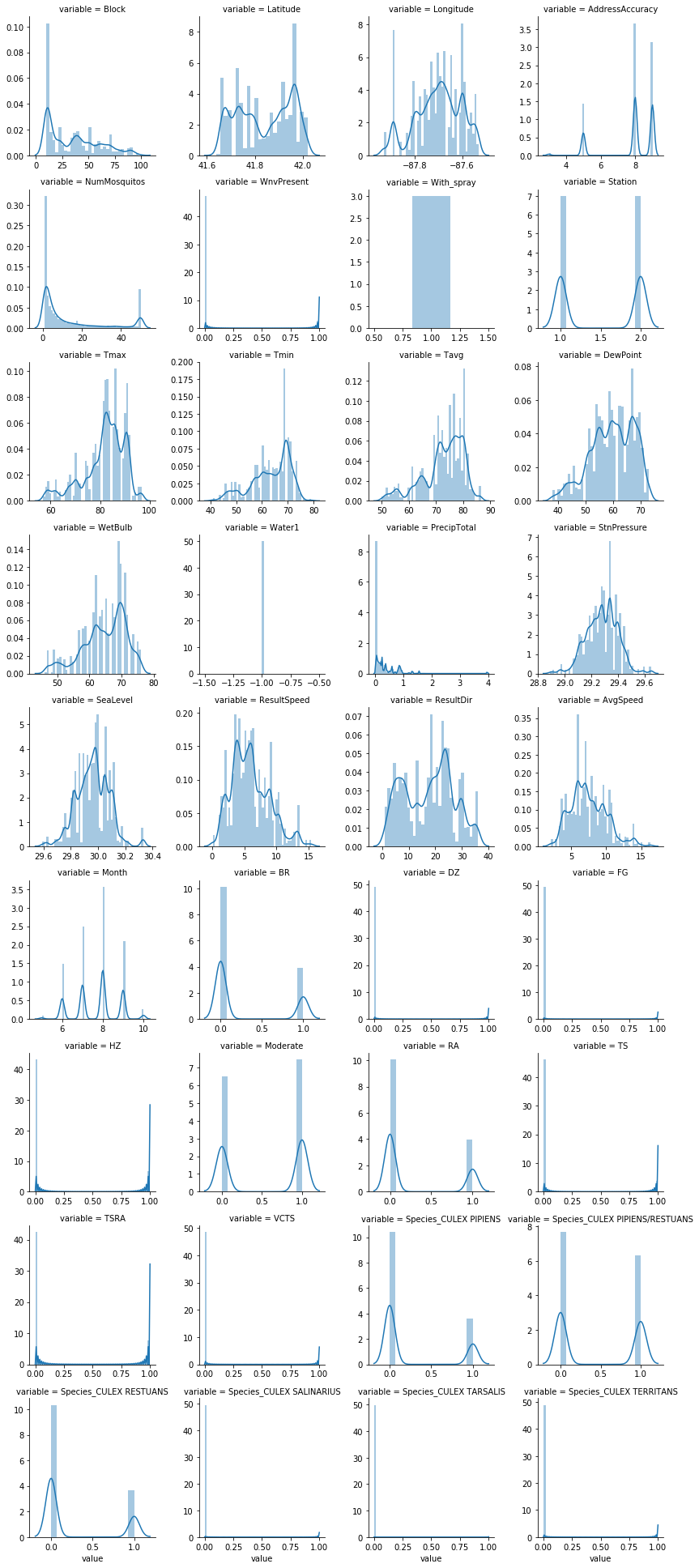
# see how is number of Mosquitos like over time
fig = plt.figure(figsize=(15,30))
ax1 = fig.add_subplot(211)
ax1 = sns.countplot(x='mm', data=combined_data, color='darkgreen', order=sorted(unique_date))
ax1 = plt.xticks(rotation=90)
ax1 = plt.xlabel('Dates', fontsize=15)
ax1 = plt.ylabel('Mosquito Count', fontsize=15)
ax1 = plt.title('Mosquito over Time', fontsize=20)

# facetgrid on the continuous features
continuous_feat = combined_data.select_dtypes('number')
f = pd.melt(continuous_feat)
contg = sns.FacetGrid(f, col='variable', col_wrap=4, sharex=False, sharey=False)
contg = contg.map(sns.distplot, 'value')
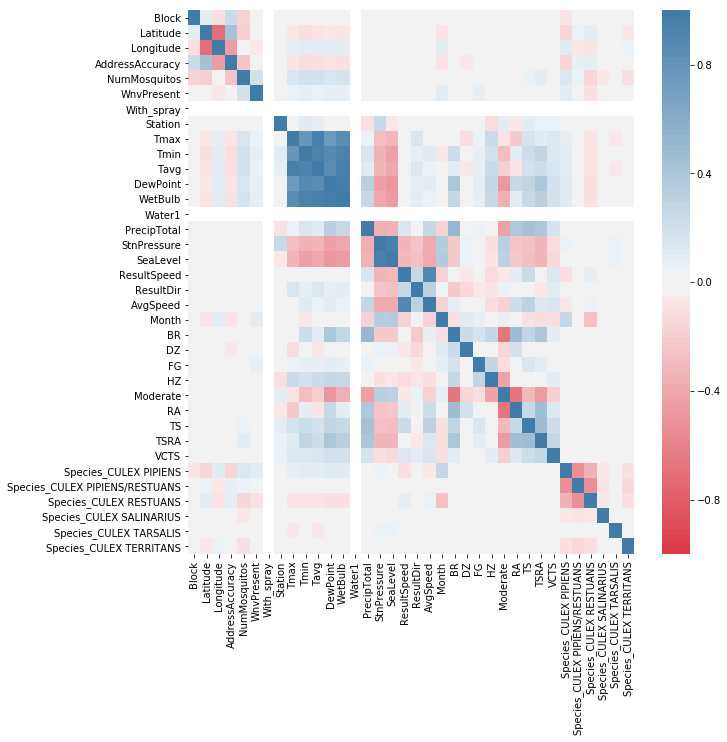
# correlation of the features and target
plt.figure(figsize=(10,10))
sns.heatmap(combined_data.corr(), cmap=sns.diverging_palette(10, 240, n=105), vmin=-1, vmax=1)
plt.show()
Modeling
# select some of the features and target
feature = ['Latitude', 'Longitude', 'Station', 'Tavg', 'DewPoint', 'WetBulb', 'PrecipTotal', 'StnPressure', 'AvgSpeed', 'Month', 'BR', 'DZ', 'FG', 'HZ', 'Moderate', 'RA', 'TS', 'TSRA', 'VCTS', 'Species_CULEX PIPIENS', 'Species_CULEX PIPIENS/RESTUANS', 'Species_CULEX RESTUANS',
'Species_CULEX SALINARIUS', 'Species_CULEX TARSALIS', 'Species_CULEX TERRITANS']
target = combined_data['WnvPresent']
# split into train and test set (hold out 33%)
X_train, X_test, y_train, y_test = train_test_split(combined_data[feature], target, test_size=0.33, random_state=42)
Train X: (14083, 25)
Train Y: (14083,)
Test X: (6937, 25)
Test Y: (6937,)
# when working with data, always take note of imbalance class as it will affect your model's accuracy
y_train.value_counts()
# Woah, quite a severe imbalance class
0 13334
1 749
Name: WnvPresent, dtype: int64
I am going to make use of SMOTETomek (imbalanced-learn package) to deal with the imbalance class. SMOTE is an up-sampling method, while Tomek Links is a down-sampling method.
# apply smote and tomek link to upsample the positive virus set
smt = SMOTETomek(n_jobs=-1)
X_resampled, y_resampled = smt.fit_sample(X_train, y_train)
Counter(y_resampled)
Counter({0: 13002, 1: 13002})
It is always good practice to scale your data so that all the features values are brought to a common degree of magnitude.
# scale the features
ss = StandardScaler()
ss.fit(X_resampled)
Xs = ss.transform(X_resampled)
Will be using classification models Logistic Regression (Baseline), Random Forest
# Logistic Regression
lr = LogisticRegression()
lr_cvs = cross_val_score(lr, Xs, y_resampled, cv=5, n_jobs=-1, scoring='recall')
print('Logistic Regression Model')
print(lr_cvs)
print('Mean Recall Score', np.mean(lr_cvs))
Logistic Regression Model
[0.77201077 0.77201077 0.77230769 0.76307692 0.79692308]
Mean Recall Score 0.7752658444976783
# check the top 8 coeff
lr.fit(Xs, y_resampled)
lr_coef = pd.DataFrame(lr.coef_, columns=X_train.columns, index=['Weight']).transpose()
print(lr_coef.sort_values(by='Weight', ascending=False).head(8))
Weight
Species_CULEX PIPIENS 1.020739
Species_CULEX PIPIENS/RESTUANS 0.866898
DewPoint 0.613928
Month 0.572167
StnPressure 0.316257
Species_CULEX RESTUANS 0.233000
Tavg 0.151438
TS 0.129305
# Random Forest
rdc = RandomForestClassifier()
rdc_cvs = cross_val_score(rdc, Xs, y_resampled, cv=5, n_jobs=-1, scoring='recall')
print('Random Forest Classifier Model')
print(rdc_cvs)
print('Mean Recall Score', np.mean(rdc_cvs))
Random Forest Classifier Model
[0.86889658 0.96962707 0.96884615 0.96538462 0.96923077]
Mean Recall Score 0.9483970366427114
# check the top 8 coeff
rdc.fit(Xs, y_resampled)
rdc_coef = pd.DataFrame(rdc.feature_importances_, index=X_train.columns, columns=['Weight'])
print(rdc_coef.sort_values(by='Weight', ascending=False).head(8))
Weight
Longitude 0.171631
Latitude 0.165150
Month 0.142626
Species_CULEX RESTUANS 0.081074
Tavg 0.081049
AvgSpeed 0.061578
DewPoint 0.050378
WetBulb 0.050120
Visualizing the Tree
I have generated a snapshot of the random forest trees, but seems like this forest is a bit too large to fit into this blog. (PSA: Don’t squint eye, you can try to generate the forest out using your own data set )

Comments
It is interesting that Random Forest prediction if a mosquito carries the West Nile virus assigns a higher weight on the location and month. In the cross validation result, the Random Forest Classifier has a higher recall score compared to the Logistic Regression baseline model. (Recall: 95% vs 78%) However, I am a little concern about the potential of over-fitting in the model when I throw in the hold out test set.
Evaluating the Model
# scaling the test set
Xtest_s = ss.transform(X_test)
# Using Logistic Regression
lr_pred = lr.predict(Xtest_s)
print(classification_report(y_test, lr_pred, labels=lr.classes_))
precision recall f1-score support
0 0.98 0.64 0.77 6584
1 0.10 0.73 0.17 353
avg / total 0.93 0.64 0.74 6937
# Confusion Matrix
lr_cm = pd.crosstab(y_test, lr_pred, rownames=['True'], colnames=['Predicted'], margins=True)
lr_cm
| Predicted | 0 | 1 | All |
|---|---|---|---|
| True | |||
| 0 | 4209 | 2375 | 6584 |
| 1 | 95 | 258 | 353 |
| All | 4304 | 2633 | 6937 |
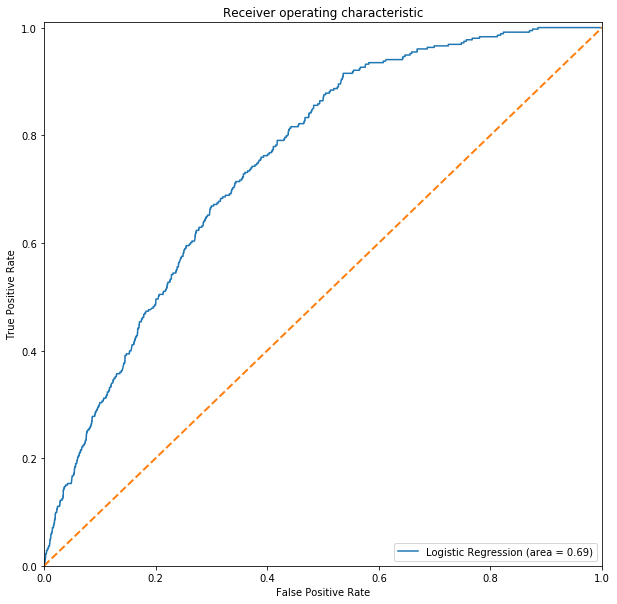
# Charting ROC-AUC
lr_roc_auc = roc_auc_score(y_test, lr_pred)
fpr, tpr, threshold = roc_curve(y_test, lr.predict_proba(Xtest_s)[:,1])
def ROC(fp, tp, roc_auc, label):
'''
fp: False Positive rate
tp: True Positive rate
label: "model name"
roc_auc: roc auc score
'''
plt.figure(figsize=(10,10))
plt.plot(fp, tp, label=label + f' (area = {roc_auc:.2f})')
plt.plot([0,1], [0,1], lw=2, linestyle='--')
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.01])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Receiver operating characteristic')
plt.legend(loc="lower right")
plt.show()
ROC(fpr, tpr, lr_roc_auc, label='Logistic Regression')
# Using Random Forest
rdc_pred = rdc.predict(Xtest_s)
print(classification_report(y_test, rdc_pred, labels=rdc.classes_))
precision recall f1-score support
0 0.96 0.92 0.94 6584
1 0.19 0.36 0.25 353
avg / total 0.92 0.89 0.90 6937
# Confusion Matrix
rdc_cm = pd.crosstab(y_test, rdc_pred, rownames=['True'], colnames=['Predicted'], margins=True)
rdc_cm
| Predicted | 0 | 1 | All |
|---|---|---|---|
| True | |||
| 0 | 6028 | 556 | 6584 |
| 1 | 226 | 127 | 353 |
| All | 6254 | 683 | 6937 |
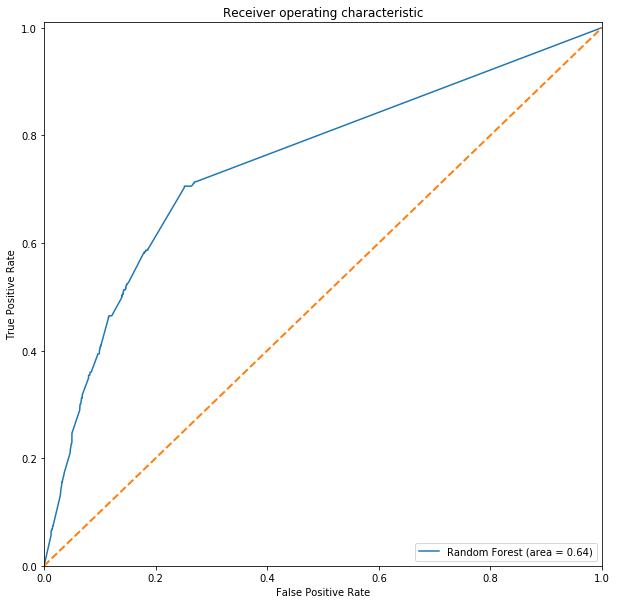
# Charting ROC-AUC
rdc_roc_auc = roc_auc_score(y_test, rdc_pred)
fpr, tpr, threshold = roc_curve(y_test, rdc.predict_proba(Xtest_s)[:,1])
ROC(fpr, tpr, rdc_roc_auc, label='Random Forest')
Observation
When the test set is thrown in for the prediction, Logistic Regression was able to generalize better and is able to pick up those mosquito with the West Nile virus accurately.
I will need to do more adjustment and perimeter tuning for the model to detect the West Nile virus.
Grid-Search
# setting up random forest param
param = {'n_estimators': [5, 10, 20],
'max_depth': [30, 50, 100],
'max_features': [2, 3],
'min_samples_split': [8, 10, 12],
'min_samples_leaf': [3, 4, 5]
}
# Gridsearch on random forest
rdc_gs = GridSearchCV(rdc, param, n_jobs=-1, cv=3, scoring='recall')
rdc_gs.fit(Xs, y_resampled)
GridSearchCV(cv=3, error_score='raise',
estimator=RandomForestClassifier(bootstrap=True, class_weight=None, criterion='gini',
max_depth=None, max_features='auto', max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=10, n_jobs=1,
oob_score=False, random_state=None, verbose=0,
warm_start=False),
fit_params=None, iid=True, n_jobs=-1,
param_grid={'n_estimators': [5, 10, 20], 'max_depth': [30, 50, 100], 'max_features': [2, 3], 'min_samples_split': [8, 10, 12], 'min_samples_leaf': [3, 4, 5]},
pre_dispatch='2*n_jobs', refit=True, return_train_score='warn',
scoring='recall', verbose=0)
# Drop out unwanted columns & show the top 10 parameter which will boost Random Forest recall
to_drop = ['mean_fit_time', 'std_fit_time', 'mean_score_time', 'std_score_time', 'params',
'split0_test_score', 'split1_test_score', 'split2_test_score', 'std_test_score',
'split0_train_score', 'split1_train_score', 'split2_train_score', 'mean_train_score', 'std_train_score']
df_rdc_gs = df_rdc_gs.drop(to_drop, axis=1)
df_rdc_gs.sort_values(by='rank_test_score').head(10)
| param_max_depth | param_max_features | param_min_samples_leaf | param_min_samples_split | param_n_estimators | mean_test_score | rank_test_score |
|---|---|---|---|---|---|---|
| 30 | 2 | 3 | 8 | 20 | 0.952930 | 1 |
| 50 | 3 | 3 | 10 | 20 | 0.951777 | 2 |
| 50 | 3 | 3 | 8 | 20 | 0.951700 | 3 |
| 100 | 3 | 3 | 8 | 20 | 0.951238 | 4 |
| 100 | 3 | 3 | 10 | 20 | 0.950777 | 5 |
| 30 | 3 | 3 | 8 | 10 | 0.950315 | 6 |
| 50 | 3 | 3 | 12 | 20 | 0.950315 | 6 |
| 50 | 3 | 3 | 8 | 10 | 0.950085 | 8 |
| 30 | 3 | 3 | 8 | 20 | 0.949931 | 9 |
| 50 | 3 | 4 | 10 | 20 | 0.949931 | 9 |
# Using best parameter from Random Forest Gridsearch to predict
rdc_gs_pred = rdc_gs.predict(Xtest_s)
print(classification_report(y_test, rdc_gs_pred, labels=rdc.classes_))
precision recall f1-score support
0 0.97 0.89 0.93 6584
1 0.18 0.47 0.26 353
avg / total 0.93 0.87 0.89 6937
# confusion matrix
rdc_gs_cm = pd.crosstab(y_test, rdc_gs_pred, rownames=['True'], colnames=['Predicted'], margins=True)
rdc_gs_cm
| Predicted | 0 | 1 | All |
|---|---|---|---|
| True | |||
| 0 | 5838 | 746 | 6584 |
| 1 | 188 | 165 | 353 |
| All | 6026 | 911 | 6937 |
# Charting ROC-AUC
rdcgs_roc_auc = roc_auc_score(y_test, rdc_gs_pred)
fpr, tpr, threshold = roc_curve(y_test, rdc_gs.predict_proba(Xtest_s)[:,1])
ROC(fpr, tpr, rdcgs_roc_auc, label='Random Forest with GridSearch')
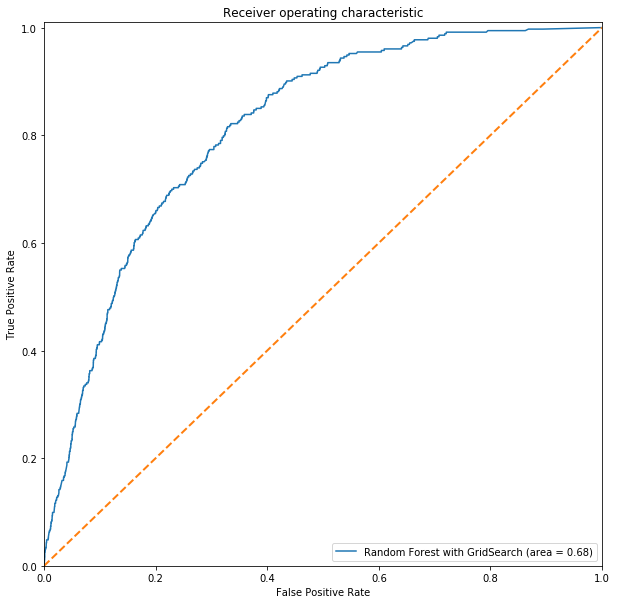
Comments
While the average recall score has dropped on the test set, Random Forest is now able to detect mosquito with the west nile virus better.
